Special Report on Microbiomics: The aliens within us
As if human diversity wasn’t tricky enough
Register for free to listen to this article
Listen with Speechify
0:00
5:00
Right now, as you read this article, you are under alien surveillance. If you lower these pages or raise your eyes from the computer screen, you may think you see your co-worker or life partner across the room, but none of these people are who they present themselves to be, or at least not fully.
Neither is your favourite child, your hockey teammate or your dog. In fact, neither is the person staring back at you from the mirror.
You’re unlikely to get a visit from any real-life version of the cinematic agents from “Men in Black,” however, nor is there is much likelihood that you will pull a John Hurt-style scene from “Alien” over the dinner table.
That said, you are more alien than human. It is the alien within us all, however, that is keeping us alive and functioning.
A walking ecosystem
Almost every nook and cranny of the human body, both inside and out, is home to myriad microorganisms—viruses, fungi, bacteria and others that comprise the microbiome. It might be more accurate, however, to suggest that the human body occupies the space in between the microbes, as those “foreign” organisms account for 90 percent of a human’s physical being and more than 99 percent of its communal genome. Granted, while they outnumber human cells around 10-to-1 on an individual basis, their small size means they only account for 1 to 3 percent of the body’s mass. Still, they pack a wallop in terms of their role in human health.
Much as the Amazonian rainforest is home to a vast, interconnected ecosystem, so too are we. And as the loss of a single species in that forest can have a devastating ripple effect, the same is also true for us when the natural balance of organisms is disrupted, shifting to dysbiosis and disease.
“I think the relationship between dysbiosis and disease is still not well defined,” offers Alexander Maue, associate director for microbiome products and services at Taconic Biosciences, a company that provides germ-free mice to facilitate these questions.
“Does dysbiosis cause disease, or is dysbiosis the result of disease?” he asks.
The answer may be a bit of both.
“There are examples where dysbiosis can lead to disease,” Maue continues. “The most important example would be that of Clostridium difficile, where antibiotic treatment disrupts the microbiome and allows C. difficile to take hold and cause disease.”
It is less clear, he says, with inflammatory bowel disease (IBD) as to whether the dysbiosis causes the disease or is the result of the disease.
“Although, the dysbiosis itself could serve to exacerbate the disease,” he adds.
Whatever the relationship, the added complexity of microbial input to everything from basic human metabolism to immunological development to response to drugs has pushed microbiomic analysis to the forefront of research into health and disease.
“The whole concept of the microbiome, I believe, is the next paradigm shift in science,” says Glenn Nedwin, CEO of Second Genome, a company hoping to leverage microbiomic insights into new biotherapies.
As big as the development of recombinant DNA technologies was to biomolecular research, Nedwin argues, providing us with the myriad biotherapeutics in use today, an expanded understanding and harnessing of the microbiome stands to be a magnitude more important.
The first step is trying to identify distinct microbes that only exist in either the healthy or diseased state.
“Let’s say you have people with cancer and only 20 percent are responding to the drug,” Nedwin explains. “You would take, let’s say, 300 patients of varying degrees of fully responsive to the drug, not at all and healthy people, and you would then profile the bacteria in all of these samples.”
Are there unique microbes in the samples of the 20 percent who respond to the drug?
“The answer is going to be yes,” he assures. “Then you have to find out what those microbes are.”
And this is the point at which things get tricky, because despite the wealth of microbiological experience in culturing bacteria, viruses and fungi, the vast majority of the microbiome is intractable to expansion by culturing.
“There used to be a lot of literature that we could only culture 1 percent of the organisms, but I think that number is quite a bit higher today; more like 25 or 30 percent,” Nedwin recounts. “And of course, these microbes are highly anaerobic.”
But even with these larger numbers, the concern remains that microbes within an ecosystem may not be identical to microbes grown in isolation. Fortunately, other technologies have stepped up to fill in the gap.
Taking a survey
“The advent of next-generation sequencing technologies has revolutionized our view of human-associated microbial communities,” explained the University of Pennsylvania’s Elizabeth Grice in a 2014 review. “Using DNA sequencing methodology, we are now able to characterize and analyze microbiomes with greater precision and accuracy, and less bias compared to culture-based approaches.”
“A common approach used to identify bacterial populations is based on sequencing of the small subunit bacterial 16S ribosomal RNA (rRNA) gene,” she continued, adding that for fungi, researchers similarly rely on the 18S and 5.8S rRNA gene sequences.
Recently, the University of Florence’s Rolando Cimaz and colleagues used this approach to examine the gut microbiota of healthy subjects and patients with different forms of juvenile idiopathic arthritis, a common chronic rheumatic disorder of children. The goal was to identify possible pro-arthritogenic microbial signatures.
Performing pyrosequencing on microbes isolated from fecal samples, the researchers noted distinct taxonomic distributions between the gut microbiomes of the different groups, some of the population changes (up or down) being reflective of previous studies in other inflammatory conditions such as Crohn’s disease or ankylosing spondylitis.
They also noted significant differences in the stability of microbiota profiles depending on the individual’s disease status.
“As expected, our results suggest that during active disease, the microbiota is strongly perturbed (major intra-group distance compared with remission),” the authors described. “Healthy status allows a more stable microbiota ecosystem compared to disease status, as previously observed.
“Remission is characterized by an intermediate microbial pattern, different from both active disease and healthy controls, likely resulting from a different trajectory to stable state and in which autoimmune reactivity and the microbial ecosystem are mutually shaped.”
Despite the awareness provided by studies such as these, however, Nedwin questions how useful they are to the ultimate characterization and treatment of the disease as such studies typically limit their microbial identification to broadly defined taxonomy. For Nedwin, strains matter.
“These massive groupings that most people publish are nothing actionable,” he argues. “You can’t say there is Bacillus in there, because there are thousands of different bacilli. We all know, for example, with E. coli, the one that makes us sick is E. coli O147, but we have a lot of E. coli which keeps us healthy.”
More broadly, Maue agrees that next-generation sequencing methodologies have been vital to helping us understand the number and diversity of microbes in the gut. For him, however, the greater information will come from the applications of technologies in areas such as metabolomics, which he thinks are essential to the development of new classes of biologic therapies and drugs.
His thoughts were recently echoed by his Taconic colleague Philip Dubé, who suggested that whereas the species diversity in a given microbiome may be relatively stable throughout the life of a host organism, the metabolic activities of those same microbes may not.
“Bacteria are highly adaptable to differing energy sources, such as diet-specific complex carbohydrates and fatty acids, and can produce divergent metabolites as their diet is altered, “ he wrote. “This raises the possibility of targeting the microbiome at its functional level for therapeutic interventions—an intriguing strategy given the apparent intractability of altering microbiome composition in a durable fashion.”
In a recent discussion of microbiomic profiling approaches, Pamela Vernocchi and colleagues at the Bambino Gesú Children’s Hospital reflected on the importance of understanding microbial metabolism in human health.
“The gut microbiota is involved in several biochemical functions directly associated to the perturbation of specific gut microbial populations, which may lead to the development of diseases,” they suggested. “Moreover, the identification of metabolites may highlight how lifestyle and dietary habits affect specific disease conditions.
“Finally, metabolomics represents an unprecedented approach to collect the complex metabolic interactions between the host and its commensal microbial partners, offering the opportunity to define individual and population phenotypes.”
Interestingly, the authors also commented on one of the broader philosophical challenges facing microbiomic studies and the use of microbiome-directed treatment: whether to take a holistic or reductionist approach.
For Vernocchi and her colleagues, it seems to be less an either/or scenario and more a question of 'why not both?'
“The combination of these [metabolomics] techniques with genome analysis may lead to a holistic view of the metabolic pathways, which can also be backed up using mathematical models and statistical assessment of data,” they explained. “In fact, by managing data, it is possible to achieve a higher level of biological understanding.
“Therefore, novel algorithms and statistical analysis need to be improved to integrate the ‘omics’ data, and a stochastic model of metabolic networks needs to be introduced to lead to a novel knowledge of co-regulation in biochemical networks.”
Another resource that is seeing increased use in microbiomics research is the germ-free mouse, which, being devoid of bacteria, is the ideal substrate for the reductionist microbiome approach, according to Maue.
In earlier studies, researchers often treated test animals with broad-spectrum antibiotics to reduce or oblate the microbiome. Maue suggests, however, that microbial removal was often incomplete, and therefore the utility of these models was limited.
With germ-free mice, however, researchers can partially or completely reconstitute the gut microbiome, allowing them to study the impacts of the various combinations on mouse metabolism and disease response.
“You can control a lot of the variability with mice because we have a comprehensive knowledge of mouse genetics,” he continues. “The mouse models themselves have a relatively low cost of maintenance. They have a high reproductive rate and short life cycle.”
And as mouse models can now be humanized with the replacement of mouse genes or tissues with their human counterparts, there is a growing opportunity to examine the impact of microbiotic changes on human immune cell development or liver metabolism, for example (see also the article “Identifying toxicity” in the October 2016 issue of DDNews).
“There are complexities, however, regarding the full reconstitution of mucosal immunity,” Maue clarifies. “To be frank, I think additional discoveries are going to be needed to untangle the discrete immunologic properties that can be modeled in the humanized immune system mouse.”
“I think that once we understand the biological underpinnings, there is going to be immense potential to examine combinations of human microbiome and human immune cell functions in vivo,” he enthuses.
And once you have a model system in which you can essentially recapitulate the human microbiotic experience, opportunities abound to then influence those disease models.
Therapeutic approaches
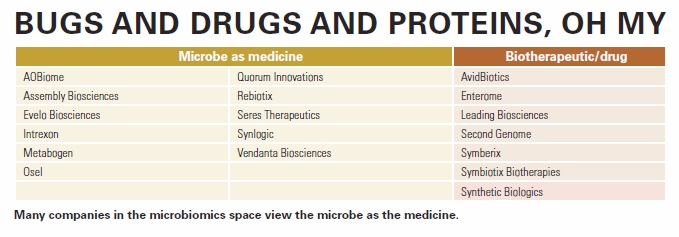
“There are different ways of approaching the microbiome,” says Nedwin, pointing out that most companies in this space view the microbe as the medicine (see also chart “Bugs and drugs and proteins, oh my” below).
The broadest approach is fecal microbiota transplant (FMT), which has shown strong efficacy in the treatment of C. difficile infection (CDI), with typical recovery rates in the 80- to 90-percent range or higher. Here, the microbes found in the stools of healthy individuals essentially out-compete C. difficile for resources, re-establishing the ecological balance in the patient’s colon.
In a recent review, however, Andreas Meisel and colleagues at Universitätsmedizin Berlin highlighted their concerns about the seeming randomness of FMT.
“The approach of targeting whole communities appears to be a shot in the dark: the outcome of such a therapy is not only unpredictable, but might also have more severe adverse effects, in particular when the entire microbial community is profoundly influenced,” they wrote. “Since the field of microbial therapy is in its infancy and little is known about side effects, transplantation of currently unknown pathogenic microorganisms by FMT cannot be excluded.”
As an example, they described the transplantation of intestinal microbes from atherosclerosis-prone mice to another strain that was disease-resistant. The transplant was sufficient to induce susceptibility to atherosclerosis.
“Thus, even before risk factors for diseases manifest gut microbiota might have pathogenic potential, making the definition of healthy persons as donors for FMT inherently difficult.”
In recognition of concerns such as these, Seres Therapeutics has taken a slightly different approach, viewing stools as simply the starting point. Using fecal samples from healthy donors, the company fractionates the material to isolate microbial spores. The company’s lead product SER-109 is comprised of approximately 50 species of Firmacutes spores and has shown strong efficacy in rodent models of CDI.
Late last year, Seres’ Barbara McGovern and colleagues presented their efforts to use SER-109 in CDI patients following standard-of-care antibiotic treatment. They noted that almost 90 percent of patients showed no C. difficile-positive diarrhea up to eight weeks post-treatment, and this rate jumped to 97 percent when including patients who resolved by eight weeks.
The researchers also monitored the impact of treatment on patient microbiomes, which they found returned to a state more similar to those found in healthy individuals.
“Seeding of the microbiome with a purified fraction of spores led to marked diversification of commensal bacteria, including augmentation of species not present in SER-109,” the authors wrote. “The engraftment and augmentation of commensals observed in recipients of SER-109 suggests that this spore-based approach provides the ecologic scaffolding needed to promote the regrowth of a diverse microbial community that is shaped by the individual host.”
The researchers also suggested that the use of fractionated spores decreased the likelihood of transmission of potentially infectious microbes that might be transmitted from asymptomatic donors, or proinflammatory macromolecules that could activate innate immunity in the recipient gut.
Rebiotix is following a similar path with its lead candidate RBX2660, which is also a fractionated product of donor stool samples. Over the past two years, the company has reported the results of its two Phase 2 trials in patients with recurrent CDI, noting efficacy rates approaching 90 percent with no unanticipated adverse events.
The company is currently working on an oral version of its recently patented Microbiota Restoration Therapy for recurrent CDI, as well other therapeutic areas such as ulcerative colitis and hepatic encephalopathy.
Other groups, meanwhile, are honing their research efforts more tightly, focusing on single microbes rather than collections.
Very recently, Veena Taneja and colleagues at University of Iowa and the Mayo Clinic described their efforts to use a single probiotic to suppress inflammatory arthritis in a humanized mouse model. After inducing arthritis via collagen immunization, the researchers then fed the mice two strains of commensal Prevotella spp. or bacterial media alone.
They found that only mice that received P. histicola showed reductions in proinflammatory cytokines and chemokines, and that the impact was the same whether the probiotic was given before immunization (prophylactic) or after (therapeutic). The reduced arthritis severity following treatment was also noted in a second mouse model, suggesting the effect was not genotype-specific.
And unlike many biologic drugs used to treat rheumatoid arthritis, the probiotic did not lead to suppression of innate immune responses in the mice, offering the hope that the commensal bacteria may be safer in some ways. Further studies of this are required, however, the authors were quick to note.
In some cases, though, rather than add something to the microbiotic mix, it might be better to remove the offending party, but with much greater specificity than is typically afforded by antibiotic treatments. For Nestlé’s Harald Brüssow and colleagues, the answer was bacteriophage, viruses that attack bacteria.
In a recent safety trial on healthy and diarrheal children from Bangladesh, the researchers treated the patients with an oral cocktail of T4-like phages, a commercial phage cocktail or placebo.
Regardless of which phage preparation was used, no significant adverse events were noted. And perhaps more importantly, the percentage of phage-positive stools in healthy subjects was proportional to oral dose, suggesting that the phage did not replicate. Of note, the researchers saw no significant treatment effect of the oral phage application versus standard treatment (placebo).
Nedwin is unconvinced that microbe as medicine is the way to go, however, suggesting that it leaves a lot of questions open regarding not just safety, but also mechanism of action.
“If you’re using the whole bacteria or groups, you need to understand what it is doing when you put it back into a person,” he says. “How is that manipulating the communities that are there now, and of the bacteria that you put in, are they tourists or tenants? Will they grab hold and start colonizing or are they just going through, which is what most probiotics do, so there is not a long-term effect.”
In 2015, Jeremy Burton, deputy director of the Canadian Centre for Human Microbiome and Probiotic Research offered much the same opinion.
“In chronic diseases, using single- or multi-strain probiotic bacterial strains is a bit like trying to recolonize the fauna of a decimated jungle with one or two domesticated animals,” he wrote in a commentary. “In reality, few probiotics ever persist after dosing and can also be quite different physiologically and genetically from their original isolates.”
He also questioned whether it was likely the clinic would ever see microbes that hadn’t already been well characterized for their roles in the food industry, such as Lactobacilli and Bifidobacteria, which he suggests are generally minor components of the intestines.
“Most intestinal bacteria which currently ‘roam free’ will never see their use as probiotics, given the regulatory and safety requirements needed to bring to market, although the probiotics market is valued as high as $30 billion.”
For Nedwin, there is also the practical consideration of manufacturing.
“When you’re talking about the fecal transplant or growing up strains, you need to grow them up individually,” he argues. “From a manufacturing standpoint, that’s a nightmare.”
“Any microbe you grow—I don’t care if it’s yeast, E. coli or whatever—there are subtle changes that are made every time you grow it,” he presses. “So, it’s not always exact 100 percent, and that difference can matter. When you put it together, it may not do what you think you are doing.”
Eschewing “bugs as drugs,” Nedwin’s Second Genome has taken a different approach, going down the more traditional road to biotherapeutics development.
The company started with a typical differential analysis of what bacteria were in healthy individuals vs. patients with disease, but rather than stop with the bacteria, they applied metagenomics, genome sequencing, bioinformatics and machine learning to dissect out what made those bacterial species critical to health or disease.
“We’re saying let’s just find the molecule that makes your epithelial cell healthy and you can take that as an oral protein,” he says, explaining that the company now has a pipeline of 14 natural biomolecules isolated from bacteria that maintain the health of gut epithelial cells.
The company’s lead product SGM-1019 is an orally delivered small-molecule inhibitor of an undisclosed target, and has completed Phase 1 studies for the treatment of IBD. The company is also developing compounds targeting metabolic disorders such as obesity and type 2 diabetes, as well as other conditions stemming out of the gut focus, such as immunological and neurological diseases.
Taking a slightly different approach, Synthetic Biologics is looking to protect the gut microbiome from systemic beta-lactam antibiotics that can lead to conditions such as CDI or antibiotic-associated diarrhea as well as trigger the emergence of resistant microbes. The company’s lead product SYN-004, or ribaxamase, is an oral enzyme that degrades the beta-lactam antibiotics as they enter the digestive tract, and is currently in Phase 2b trials. The work is supported, in part, by a research contract the company signed with the U.S. Centers for Disease Control and Prevention (CDC) in October.
“Antibiotics are lifesaving medicines, but they also can disrupt a person’s microbiome and increase the risk for drug-resistant infections,” suggested the CDC’s Clifford McDonald, associate director of science in the Division of Healthcare Quality Promotion, in the announcement. “To protect people, their microbiomes and the effectiveness of antibiotics, this project is an example of applied research that has the potential to produce innovative public health approaches to better combat antibiotic resistance.”
Leading Biosciences, meanwhile, is focusing its energies on the intestinal mucosal barrier, recently launching what it calls its gastrobiome program.
“The most important aspect of the gastrobiome is the permeability of the mucosal barrier, which traps the disease-causing elements in the small intestine while allowing essential nutrients to cross the barrier for absorption by the body,” explained Thomas Hallam, senior vice president of clinical development and regulatory affairs. “Our research has demonstrated that the barrier can become leaky, allowing small amounts of disease-causing elements through the barrier and driving chronic diseases such as diabetes and hypertension. Furthermore, the barrier can fail suddenly, like water breaking though a levee, causing a medical crisis such as multiorgan failure.”
The company’s lead product LB1148 is an oral broad-spectrum serine protease inhibitor currently in Phase 2 studies in post-operative ileus and adhesions. The product restores and maintains the mucosal barrier, thereby keeping the digestive enzymes from leaking through the intestinal walls into the surrounding tissues and organs.
Building consensus
Despite these early successes and the rapid identification of correlations between perturbations in various human microbiomes and diseases, there is still a lot of consensus to build among the results and the researchers. Questions of whether the therapeutic focus should be based on the microbe or biomolecular pathways pales in comparison to more fundamental discussions about microbiota networks and variability within and between populations.
Whereas one study suggests that a microbiome becomes relatively stable once a human subject passes through childhood, another study indicates large fluctuations in microbial populations occur in response to temporal, dietary or environmental changes. How important these fluctuations are to broader biological and pathological questions remains to be seen, and perhaps, if examined through a metabolic lens, the changes are merely cosmetic in the biochemical sense.
As the cataloguing work of microbe identification diminishes, perhaps more of these questions will be answered.
More than gut instinct
Although the gut is the predominant source of microbial habitation and diversity in the human microbiome, other tissues of the body are home to ecosystems, including mucosal membranes of the pulmonary system and genitourinary tract, as well as the skin.
It is, in fact, the microbiomes of these last two locations that dictate the formative microbiota of newborn infants essentially based on the mechanism by which they were born.
“Analysis of neonate skin microbiota indicates that those babies delivered via vaginal birth had skin microbiota similar to their mother’s vaginal microbiota, and babies delivered via Caesarean section had skin microbiota similar their mother’s skin microbiota,” explained University of Pennsylvania’s Elizabeth Grice in a 2014 review.
“It is unclear how long these colonization differences persist, as most vaginal bacteria are not typically found on the skin, and if the associated differences translate into functional consequences and/or disease risk.”
She suggested that other factors playing a role in skin microbiome composition include geographic location, lifestyle and even ethnicity. And further adding to this diversity is the nature of the skin itself, whether dry, moist or located in sebaceous areas such as the face and behind the ears, where lipophilic bacteria are associated with conditions such as acne and rosacea.
Areas outside of the gut are anticipated to see growing interest in microbiomic research as many of the lessons learned in early-phase research spread to other sites around the body. Although microbiomics company Second Genome has focused most of its R&D efforts on the gut, for example, it also has a microbe profiling service arm that is working with clients in several of these other areas.
Likewise, germ-free mice from companies like Taconic offer researchers unique opportunities to study the impact of specific microbes or microbe populations in mouse models of different human diseases or even within the context of mice with humanized immune or organ systems.
And the skin offers accessibility benefits that few other tissues can to microbiomic research.
“Similar to fecal microbiota transplantation therapies to treat Clostridium difficile infections, one could envision the implementation of skin microbiota transplantations to ameliorate S. aureus colonization and AD [atopic dermatitis] symptoms without relying on antibiotic therapies that encourage acquisition of resistance,” Grice suggested.
As though in response to this comment, Sandip Datta and colleagues at the National Institute of Allergy and Infectious Diseases recently described their efforts to transplant human skin microbiota into in-vitro and in-vivo models of AD, looking at culturable bacteria from healthy volunteers and AD patients.
Examining the 16S rRNA signatures of the different sources, they identified distinct microbial populations between the two groups, and these distinctions seemed to have a direct impact on S. aureus growth. Specifically, culturable gram-negative (CGN) bacteria from healthy volunteers inhibited S. aureus growth by almost 50 percent in vitro, whereas most CGN strains from AD patients failed to do so.
They then applied specific CGN strains to an induced mouse model of AD-like dermatitis and noted that R. mucosa from healthy volunteers ameliorated the condition, while that from AD patients did not, offering some hope for a topical microbial formulation. The effect was only seen in live cultures, however, as efficacy disappeared upon killing of R. mucosa from healthy individuals.

